Sentieon●体细胞变异检测-系列1
Sentieon 致力于解决生物信息数据分析中的速度与准确度瓶颈,通过算法的深度优化和企业级的软件工程,大幅度提升NGS数据处理的效率、准确度和可靠性。
针对体细胞变异检测,Sentieon软件提供两个模块:TNscope和TNhaplotyer2。
TNscope:此模块使用Sentieon特有的算法,拥有更快的计算速度和更高的计算精度,对临床基因诊断样本尤其适用;
TNhaplotyper2:此模块匹配Mutect2(现在匹配到4.1.9)结果的同时,计算速度提升10倍以上。
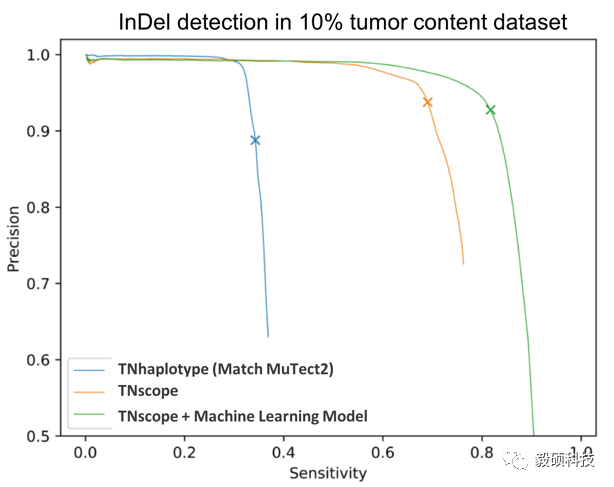
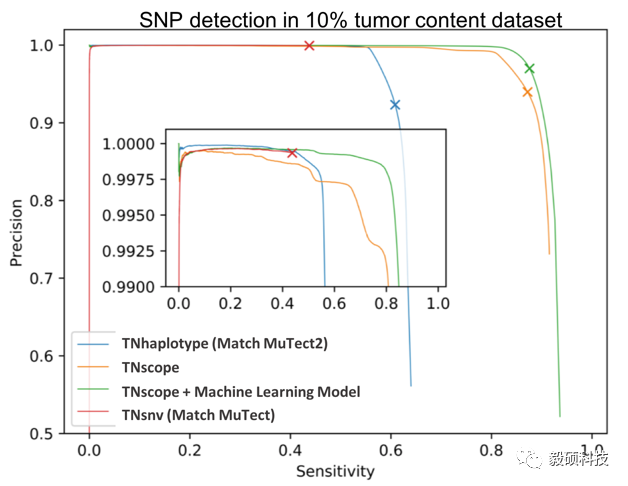
WES or Panel 变异检测分析
以下给出的步骤脚本主要针对WES or Panel (200~500x depth, AF > 1%)。(查看脚本时,可以左右滑动)
第一步:Alignment
# ****************************************** # 1a. Mapping reads with BWA-MEM, sorting for tumor sample # ****************************************** ( sentieon bwa mem -M -R "@RG\tID:$tumor\tSM:$tumor\tPL:$platform" \ -t $nt -K 10000000 $fasta $tumor_fastq_1 $tumor_fastq_2 || \ echo -n 'error' ) | \ sentieon util sort -o tumor_sorted.bam -t $nt --sam2bam -i - # ****************************************** # 1b. Mapping reads with BWA-MEM, sorting for normal sample # ****************************************** ( sentieon bwa mem -M -R "@RG\tID:$normal\tSM:$normal\tPL:$platform" \ -t $nt -K 10000000 $fasta $normal_fastq_1 $normal_fastq_2 || echo -n 'error' ) | \ sentieon util sort -o normal_sorted.bam -t $nt --sam2bam -i -
第二步:PCR Duplicate Removal (Skip For Amplicon)
# ****************************************** # 2a. Remove duplicate reads for tumor sample. # ****************************************** # ****************************************** sentieon driver -t $nt -i tumor_sorted.bam \ --algo LocusCollector \ --fun score_info \ tumor_score.txt sentieon driver -t $nt -i tumor_sorted.bam \ --algo Dedup \ --score_info tumor_score.txt \ --metrics tumor_dedup_metrics.txt \ tumor_deduped.bam # ****************************************** # 2b. Remove duplicate reads for normal sample. # ****************************************** sentieon driver -t $nt -i normal_sorted.bam \ --algo LocusCollector \ --fun score_info \ normal_score.txt sentieon driver -t $nt -i normal_sorted.bam \ --algo Dedup \ --score_info normal_score.txt \ --metrics normal_dedup_metrics.txt \ normal_deduped.bam
第三步: Base Quality Score Recalibration (Skip For Small Panel)
# ****************************************** # 3a. Base recalibration for tumor sample # ****************************************** sentieon driver -r $fasta -t $nt -i tumor_deduped.bam --interval $BED \ --algo QualCal \ -k $dbsnp \ -k $known_Mills_indels \ -k $known_1000G_indels \ tumor_recal_data.table # ****************************************** # 3b. Base recalibration for normal sample # ****************************************** sentieon driver -r $fasta -t $nt -i normal_deduped.bam --interval $BED \ --algo QualCal \ -k $dbsnp \ -k $known_Mills_indels \ -k $known_1000G_indels \ normal_recal_data.table
第四步:Variant Calling
sentieon driver -r $fasta -t $nt -i tumor_deduped.bam -i normal_deduped.bam --interval $BED -interval_padding 10\ --algo TNscope \ --tumor_sample $TUMOR_SM \ --normal_sample $NORMAL_SM \ --dbsnp $dbsnp \ --sv_mask_ext 10 \ --max_fisher_pv_active 0.05 \ --min_tumor_allele_frac 0.01 \ --filter_t_alt_frac 0.01 \ --max_normal_alt_frac 0.005 \ --max_normal_alt_qsum 200 \ --max_normal_alt_cnt 5 \ --assemble_mode 4 \ [--pon panel_of_normal.vcf \] output_tnscope.pre_filter.vcf.gz
第五步:Variant Filtration
bcftools annotate -x "FILTER/triallelic_site" output_tnscope.pre_filter.vcf.gz | \ bcftools filter -m + -s "insignificant" -e "(PV>0.25 && PV2>0.25)" | \ bcftools filter -m + -s "insignificant" -e "(INFO/STR == 1 && PV>0.05)" | \ bcftools filter -m + -s "orientation_bias" -e "FMT/FOXOG[0] == 1" | \ bcftools filter -m + -s "strand_bias" -e "SOR > 3" | \ bcftools filter -m + -s "low_qual" -e "QUAL < 20" | \ bcftools filter -m + -s "short_tandem_repeat" -e "RPA[0]>=10" | \ bcftools filter -m + -s "noisy_region" -e "ECNT>5" | \ bcftools filter -m + -s "read_pos_bias" -e "FMT/ReadPosRankSumPS[0] < -8" | \ bcftools norm -f $fasta -m +any | \ sentieon util vcfconvert - output_tnscope.filtered.vcf.gz