1. 脚本主要内容
* 批量读取下机数据
* 计算双细胞比例
* BBKNN去除批次效应
* 去除细胞周期的影响
* 转换为seurat对象
2. 脚本
点击查看代码
import scanpy as sc
import anndata as an
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import scrublet as scr
import scipy.io
import numpy as np
import os
import pandas as pd
from math import sqrt
import bbknn
import random
from matplotlib.pyplot import rc_context
import datetime
import DIY
from DIY import get_tsne
sc.logging.print_versions()
sc.set_figure_params(facecolor="white", figsize=(8, 8))
sc.settings.verbosity = 3
random.seed(234)
figure_out = './figures'
results_file = 'write/sce.h5ad'
table_out = './write'
datadir='./CellrangerOut/'
# df=pd.read_csv('sample_info.txt',header=0,index_col=0,sep='\t')
sample_type={}
samplelist = []
def InputData(SampleInfo):
metadata = pd.read_csv(SampleInfo,header=0,index_col=0,sep='\t')
filenames = metadata.index
adatas = [sc.read_10x_mtx(datadir + filename+ '/filtered_feature_bc_matrix',cache=True) for filename in filenames] # use list to store separate adata
for i in range(len(adatas)):
adatas[i].obs['sample'] = metadata.index[i]
for col in metadata.columns:
adatas[i].obs[col] = metadata[col][i]
adata = adatas[0].concatenate(adatas[1:],batch_categories = metadata.index)
adata.var_names_make_unique()
sc.pl.highest_expr_genes(adata, n_top=20, save = True )
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
# ribo_gene_df = pd.read_csv(r'KEGG_RIBOSOME.v2023.1.Hs.csv', sep=',')
# ribo_gene_df = ribo_gene_df[ribo_gene_df.STANDARD_NAME=='GENE_SYMBOLS']
# ribo_gene_names = ribo_gene_df.loc[:, 'KEGG_RIBOSOME'].values[0].split(',')
# adata.var['ribo'] = adata.var.index.isin(ribo_gene_names)
adata.var['mito'] = adata.var.index.str.startswith(('MT-', 'mt-', 'Mt-'))
sc.pp.calculate_qc_metrics(adata,
expr_type='counts', # default value
var_type='genes', # default value
qc_vars=['mito'],
percent_top=None,
log1p=False,
inplace=True)
sc.pl.violin(adata, ['n_genes_by_counts', 'total_counts', 'pct_counts_mito'],
jitter=0.4, multi_panel=True,save=True)
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(10, 5))
plt.subplots_adjust(wspace=.5)
sc.pl.scatter(adata, x='total_counts', y='pct_counts_mito',show=False,ax=ax1)
sc.pl.scatter(adata, x='total_counts', y='n_genes_by_counts',show=False,ax=ax2)
fig.savefig(os.path.join(figure_out, 'scatter.pdf'),
dpi=300, bbox_inches='tight')
return adata
# 计算doublet
sim_doublet_ratio = 2
def Compute_Doublet(adata,rate):
counts_matrix = adata.to_df()
n_cells = adata.shape[0]
scrub = scr.Scrublet(counts_matrix, expected_doublet_rate=rate,
n_neighbors = round(0.5 * sqrt(n_cells)),
sim_doublet_ratio = sim_doublet_ratio)
### annoy-1.15.1
doublet_scores, predicted_doublets = scrub.scrub_doublets(min_counts=2,
min_cells=3,
min_gene_variability_pctl=85,
n_prin_comps=30)
scrub.plot_histogram()
plt.savefig(os.path.join(figure_out,"histogram.pdf"),dpi=300, bbox_inches='tight')
plt.show()
scrub.set_embedding('TSNE', scr.get_tsne(scrub.manifold_obs_, 0.5,10))
scrub.plot_embedding('TSNE', order_points=True)
plt.savefig(os.path.join(figure_out,"predicted_doublets.pdf"))
plt.show()
out_df = pd.DataFrame()
out_df['barcodes'] = counts_matrix.index
out_df['doublet_scores'] = doublet_scores
out_df['predicted_doublets'] = predicted_doublets
#out_df.to_csv('doublet.txt', index=False,header=True)
out_df.to_csv(os.path.join(table_out,'doublet.txt'), index=False,header=True)
out_df.head()
return out_df,doublet_scores,predicted_doublets
def Filter_cells(adata,doublet_scores,predicted_doublets,mito):
adata.obs["doublet_scores"] = doublet_scores
adata.obs["predicted_doublets"] = predicted_doublets
#~ 可以作为取反的功能
adata = adata[~adata.obs.predicted_doublets, :]
upper_limit = np.quantile(adata.obs.n_genes_by_counts, 0.98)
adata = adata[adata.obs.n_genes_by_counts < upper_limit, :]
adata = adata[adata.obs.pct_counts_mito < mito, :]
return adata,upper_limit
def CellCycleScoring(adata,cycle,species):
data = pd.read_csv(cycle,sep=',',header='infer',usecols=[1,2,3,4])
if species == 'hsa':
s_genes = list(data.loc[ : ,'hsa_s.genes'])
g2m_genes = list(data.loc[ : ,'hsa_g2m.genes'])
else:
s_genes = list(data.loc[ : ,'mmu_s.genes'])
g2m_genes = list(data.loc[ : ,'mmu_g2m.genes'])
cell_cycle_genes = s_genes + g2m_genes
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.scale(adata,zero_center=False)
sc.tl.score_genes_cell_cycle(adata, s_genes=s_genes, g2m_genes=g2m_genes)
cell_cycle_genes = [x for x in cell_cycle_genes if x in adata.var_names]
adata_cc_genes = adata[:, cell_cycle_genes]
sc.tl.pca(adata_cc_genes)
sc.pl.pca_scatter(adata_cc_genes, color='phase',save='.Cycle.pdf')
adata.obs['Difference'] = adata.obs['S_score'] - adata.obs['G2M_score']
adata.write('./write/sce_raw.h5ad')
return adata
def Run_Normalization(adata,n_neighbors,n_pcs,resolution):
adata.layers['count'] = adata.X.copy()
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata,
n_top_genes=3000,
flavor='seurat',
subset=False,
batch_key='sample')
sc.pl.highly_variable_genes(adata,save=True)
adata = adata[:, adata.var.highly_variable]
sc.pp.regress_out(adata, ['Difference'])
#scale数据
sc.pp.scale(adata, max_value=10)
sc.tl.pca(adata)
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=n_pcs)
sc.tl.umap(adata)
sc.tl.leiden(adata, resolution=resolution, key_added='leiden')
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(15, 5))
plt.subplots_adjust(wspace=.5)
sc.pl.umap(adata, color=['sample'], frameon=False, ax=ax1, show=False)
sc.pl.umap(adata, color=['leiden'], frameon=False, legend_loc='on data', ax=ax2, show=False)
plt.savefig(os.path.join(figure_out,"umap.pdf"))
plt.show()
return adata
def Run_BBKNN(adata,n_neighbors,n_pcs,resolution):
sc.tl.pca(adata)
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=n_pcs)
sc.external.pp.bbknn(adata, batch_key='sample')
sc.tl.umap(adata)
sc.tl.leiden(adata, resolution=resolution, key_added='leiden')
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(15, 5))
plt.subplots_adjust(wspace=.5)
sc.pl.umap(adata, color=['sample'], frameon=False, ax=ax1, show=False)
sc.pl.umap(adata, color=['leiden'], frameon=False, legend_loc='on data', ax=ax2, show=False)
plt.savefig(os.path.join(figure_out,"umap-BBKNN.pdf"))
plt.show()
with rc_context({'figure.figsize': (5, 5)}):
sc.pl.umap(adata, color='leiden', add_outline=True, legend_loc='on data',
legend_fontsize=12, legend_fontoutline=2,frameon=False,
title='clustering of cells', palette='Set1',save ='-BBKNN-re.pdf')
adata.write(results_file)
return adata
def Show_Markers(adata):
ax = sc.pl.correlation_matrix(adata, 'leiden', figsize=(5,3.5),save= True)
adata_raw = sc.read_h5ad('./write/sce_raw.h5ad')
adata_raw.obs = adata.obs
adata_raw.obsm['X_umap'] = adata.obsm['X_umap']
adata_raw.obsm['seurat_clusters'] = adata.obsm['leiden']
adata_raw.obsm['nCount_RNA'] = adata.obsm['total_counts']
adata_raw.obsm['nFeature_RNA'] = adata.obsm['n_genes']
adata_raw.obsm['percent.mt'] = adata.obsm['pct_counts_mito']
adata_raw.write('./write/sce_raw.h5ad')
adata = adata_raw
adata.layers['count'] = adata.X.copy()
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.settings.verbosity=2
sc.tl.rank_genes_groups(adata, groupby='leiden', method='wilcoxon')
markers = sc.get.rank_genes_groups_df(adata, group=None, pval_cutoff=.05, log2fc_min=.25)
markers.to_csv(os.path.join(table_out,'all_markers.csv'), index=False,header=True)
top5 = pd.DataFrame(adata.uns['rank_genes_groups']['names']).head(5)
top5.to_csv(os.path.join(table_out,'top5_markers.csv'),index=False,header =True)
fig=plt.figure(figsize=(6,24),dpi=100)
for i in top5.columns:
plt.subplot(2, 7, int(i)+1) #做一个3*3的图 range(9)从0开始,因需要从1开始,所以i+1
sc.pl.rank_genes_groups_violin(adata, groups=str(i), n_genes=5,show=False)
plt.tight_layout()
plt.axis = 'off' #关闭坐标 让图更美观
plt.savefig(os.path.join(figure_out,"top5-markers.png"))
plt.show()
adata.layers['scaled'] = sc.pp.scale(adata, copy=True).X
sc.tl.rank_genes_groups(adata, groupby='leiden', method='wilcoxon')
sc.pl.rank_genes_groups_matrixplot(adata, n_genes=3, use_raw=False, vmin=-3, vmax=3, cmap='bwr', layer='scaled',save =True)
sc.pl.rank_genes_groups_stacked_violin(adata, n_genes=3, cmap='bwr',save = True)
sc.pl.rank_genes_groups_dotplot(adata, n_genes=3, values_to_plot='logfoldchanges', min_logfoldchange=3, vmax=7, vmin=-7, cmap='bwr',save = True)
sc.pl.rank_genes_groups_heatmap(adata, n_genes=10, use_raw=False, swap_axes=True, show_gene_labels=False,
vmin=-3, vmax=3, cmap='bwr',save =True)
os.system('/PERSONALBIO/work/singlecell/s01/.conda/envs/py4/bin/Rscript ./sceasy.R')
return adata
if __name__ == '__main__':
start = datetime.datetime.now()
SampleInfo = './sample_info.txt'
cycle = './cycle.gene.csv'
species = 'hsa' ## or mmu
adata = InputData(SampleInfo)
out_df,doublet_scores,predicted_doublets = Compute_Doublet(adata,0.06) # 双细胞比率 默认0.06
adata,upper_limit = Filter_cells(adata,doublet_scores,predicted_doublets,10) # 分辨率
adata = CellCycleScoring(adata,cycle,species)
adata = Run_Normalization(adata,50,50,0.99) # neibor pc resolution
adata = Run_BBKNN(adata,50,50,0.99)
Show_Markers(adata)
end = datetime.datetime.now()
print("程序运行时间:"+str((end-start).seconds/3600)+"h")
3. 主要结果展示
-
去除批次作用前
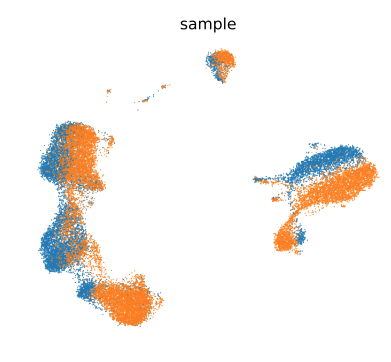
-
去除批次作用后
